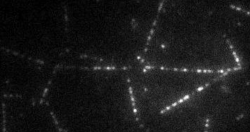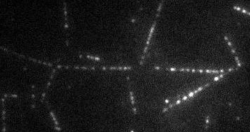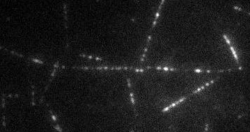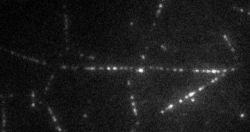
The transport of organelles and proteins is of vital importance to living cells. For these large cellular components, diffusion alone would be too slow to guarantee supply of nutrition and disposal of waste. Therefore, active transport by molecular motors is required. This transport occurs by motor proteins that walk along the cytoskeleton of the cell. Together with the group of Prof. E. Peterman (Vrije Universiteit Amsterdam), we study this cellular traffic using in vitro motility assays and Totally Internal Reflection Microscopy (TIRF). Latter provides us with distributions of fluorescently labelled molecular motors along the microtubules, see movie above. To quantify the biological traffic flow, we have developed an analysis technique yielding average velocities and run lengths of the motor proteins along the microtubule. We do this by correlating the imaged fluorescence intensities in space and time, thus following typical patterns of the distribution, while the individual motors cannot be resolved at the relevant high densities. This technique allows a quicker analysis and requires less data than the single-particle tracking approach used at low density. Moreover, correlating intensities allows analysis of data obtained under conditions inaccessible for single-particle tracking, including those performed at high motor densities, which could provide insights into traffic-jam-like interactions between motors. We find that different motor types show very different characteristics: some motor proteins keep larger distance, while others walk together in “trains”, suggesting that the cell has developed different strategies to ensure efficient traffic flow.
For more details, check out our paper in Phys. Rev. X (2017)